
Environmental DNA Metabarcoding in the San Francisco Estuary
Environmental DNA Metabarcoding in the San Francisco Estuary
Multi-year, multi-institution collaborative project funded by CA Department of Fish and Wildlife and CA Proposition 1
Project Summary
Environmental DNA (eDNA) refers to DNA originating from skin/scales, mucus, gametes, or feces that can be isolated from water and used to detect target species or reconstruct whole communities (metabarcoding) (Figure 1). In this project we are developing an eDNA metabarcoding protocol to complement existing monitoring surveys. We are creating a custom reference DNA sequence database for native and invasive fish, mussels, and macroinvertebrates present in the San Francisco Estuary (SFE). We will optimize a molecular and computational pipeline for eDNA metabarcoding and groundtruth the method against three SFE monitoring efforts, each using different sampling gear. We will investigate the relationship between eDNA sequence read count and biomass or abundance. Finally, we will determine the ability of metabarcoding to detect fish and macroinvertebrate assemblages across large and small spatial scales and over time. Our overarching goal is to develop a non-invasive, low cost monitoring tool that can be used in conjunction with existing monitoring programs or used alone to assess biological community composition at locations of interest in the SFE.
Project Significance
This project will allow more accurate and comprehensive biomonitoring and will enable better science-based decision making for managers. It will also promote increased scientific understanding of the SFE ecosystem. Multiple conservation benefits are expected to result from completion of this project. An important example is the ability to monitor whole communities, from individual water samples, that will allow managers to detect and potentially mitigate rapid shifts in SFE biological communities that are predicted to occur as the climate warms.
Objectives
Objective 1: Develop robust molecular methods and a computational pipeline for detection of SFE fish and macroinvertebrates by eDNA metabarcoding of water samples. Furthermore, we will build a DNA reference sequence database for SFE fishes, mussels and other invertebrates for this and future studies, and will create a metabarcoding protocol that will enable detection of these organisms from aquatic eDNA samples.
Objective 2: Compare eDNA metabarcoding head-to-head with existing and historical monitoring data from three ongoing ecological surveys using diverse conventional sampling gear and evaluate the accuracy of fish abundance and biomass estimates from eDNA metabarcoding data. These surveys are the enhanced delta smelt monitoring survey (EDSM), the DWR Yolo Bypass rotary screw trap, and the UC Davis Suisun Marsh Fish Study otter trawl (Figure 2). We will determine the value of eDNA metabarcoding as a survey technique by comparing species detections between it and conventional gears. We will determine whether eDNA copy/read number is positively correlated with fish species’ abundance/biomass (EDSM survey only).
Objective 3: Evaluate factors that influence eDNA detection of species of interest (e.g. rare or invasive species) and suites of species (e.g. benthic fishes and invertebrates) on two spatial scales, within and between habitats, along with temporal variation. We will determine if fine-scale spatial sampling provides higher detection rates for rare or invasive species; we will determine if surface sampling effectively detects benthic species and if benthic sampling effectively detects pelagic species. As the UC Davis Suisun Marsh Fish Study otter trawl detects fishes and some macroinvertebrates, metabarcoding results from these eDNA samples will be compared to otter trawl data to assess (presence/absence) the accuracy of both fish and invertebrate detections.
Figure 1 eDNA metabarcoding complements other monitoring methods by detecting DNA shed into the water from different aquatic species.
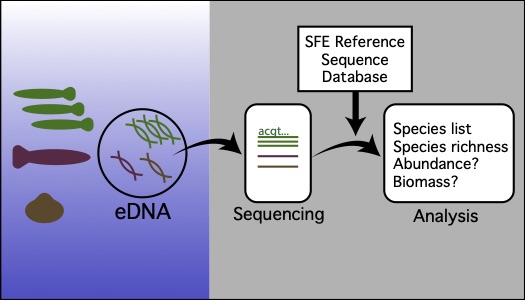
Figure 2 Project study area, including Suisun Marsh and regions currently monitored by the Yolo Bypass Fish Monitoring Program and the Enhanced Delta Smelt Monitoring Program (EDSM).

Project Team:
Andrea Schreier, Principal Investigator, Adjunct Assistant Professor, University of California, Davis
Ravi Nagarajan, Assistant Project Scientist, University of California, Davis
Ann Holmes, Graduate Student, University of California, Davis
Melinda Baerwald, Environmental Program Manager, California Department of Water Resources
Brian Schreier, Senior Environmental Scientist, California Department of Water Resources
Andrew Rypel, Associate Professor, University of California, Davis
Larry Brown, Research Biologist, US Geological Survey, California Water Science Center
Mallory Bedwell, Environmental Scientist, California Department of Water Resources
Alisha Goodbla, Lab Manager, University of California, Davis
Project Collaborators:
CA Department of Fish and Wildlife: Martha Volkoff, Heather Perry, Ian Ralston, Chelsea Callahan, Jim Snider
UC Davis: Ted Grosholz, Jessica Weidenfeld
CA Department of Water Resources: Betsy Wells, Jesse Adams
US Fish and Wildlife Service (Delta Juvenile Fish Monitoring Program): Catherine Johnston, Brian Mahardja